
J. David Lambert
- Professor
PhD
Research Active
Now accepting:
Please email with inquiries.
Please email with inquiries.
- Office Location
- 344 Hutchison
- Telephone
- (585) 273-2482
- Web Address
- Website
Office Hours: By appointment
Research Overview
We study the cellular and molecular mechanisms underlying animal development, and how these mechanisms can influence evolutionary change. We focus on embryos of molluscs and related animals, because of their large accessible cells, invariant cleavage patterns, informative phylogenetic position in the animal kingdom, and (of course!) their intrinsic beauty. Molluscs are representatives of a large clade of protostome phyla--the Lophotrochozoa--where molecular mechanisms of development are poorly understood. In addition, molluscs typify spiral cleavage, a dominant mode of early development in multiple protostome phyla and a developmental trait with an interesting evolutionary history. These embryos are amenable to a variety of classical embryological manipulations, allowing functional tests of hypotheses about patterning mechanisms. We are pursuing questions about several aspects of early patterning in the snail Ilyanassa, from the mechanisms of asymmetric cell division to signaling by the molluscan embryonic organizer. Our findings are providing new insights into the diversity of developmental mechanisms found in animals, as well as a clearer picture of how such processes evolve.
The centrosome in asymmetric cell division
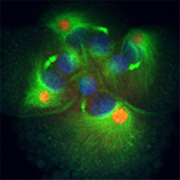
mRNAs for developmental patterning genes are localized to specific centrosomes during early cleavages in Ilyanassa. Eight cell embryo, stained for DNA (blue), microtubules (green) and IoDpp mRNA (red).
Early cleavages in spiralian embryos are often asymmetric, so that the daughter cells of a division differ in size and developmental fate. We discovered a novel mechanism for segregation of patterning molecules during asymmetric cell divisions. mRNAs of multiple developmental patterning genes are localized to particular centrosomes before they are segregated to specific daughter cells during division. We think this mechanism patterns the animal-vegetal axis of the embryo, by determining the developmental potential of tiers of cells as they are born. Many questions remain about this phenomenon, including what proportion and classes of mRNAs are localized to centrosomes, the mechanism of localization, and the developmental roles of localized patterning molecules.
Cell signaling and the dorsal-ventral axis
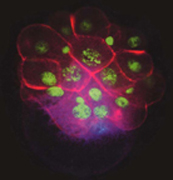
MAPK activation in the 3D nucleus (blue, lower right) precedes signaling from that cell to overlying cells to specify cell fates along the dorsal-ventral axis. Cell boundaries are red, and nuclei are green.
Classical embryological experiments have indicated that normal patterning in spiralian embryos requires induction of multiple cell fates by a cell known as the organizer. We previously demonstrated that this organizer signals via the MAPK pathway. We now have evidence that there is a parallel signal from the organizer mediated by the Dpp/BMP2&4 pathway. This signal appears to pattern the head, including positioning the eyes. This work has implications for the evolution of animal body plans. The axis inversion hypothesis holds that there has been an actual inversion in the dorsal-ventral axis during animal evolution, and the roles of Dpp/BMP2&4 pathway in flies and vertebrates have been used to support this. The role of the pathway in Lophotrochozoans is unknown and will be a key test of this hypothesis.
The evolution of spiralian patterning
We have begun to extend our work in Ilyanassa to other molluscs and related phyla. In three other molluscs and one annelid, MAPK signaling is involved in DV axis specification and organizer function, but differs significantly from its role in Ilyanassa. This is the first known commonality in the molecular mechanisms of patterning among spiralian embryos from different phyla. We will continue to examine the distribution of patterning mechanisms across spiralian and lophophorate taxa.
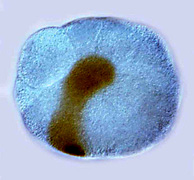
Far left: MAPK activation (brown) in the organizer cell of a chiton embryo.
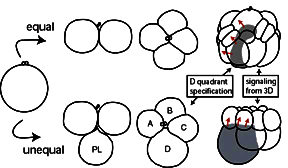
Left: the specification of the D quadrant in spiralians can occur by cellular interactions as in equal cleavers, or by asymmetric cell division as in unequal cleavers.
Genomic analysis of gene expression evolution in Drosophila sechellia
The evolution of gene regulatory activities is predicted to be an important mode of phenotypic evolution. We have been working on a project that is examining the functional relevance of gene expression evolution. This project allows genetic and genomic analyses and represents a possible bridge between microevolutionary processes and the evolution of a large-scale phenotypic change. We are studying the resistance of the fruit fly Drosophila sechellia to a plant toxin that is lethal for all related flies. The toxin, octanoic acid, is a major component of the fruit of Morinda citrifolia, and the evolution of resistance mechanisms has apparently allowed D. sechellia to occupy a new ecological niche. Unlike its closest relatives that are generalists, D. sechellia has specialized on the Morinda fruit, where it prefers to mate and lay its eggs. We are using this system, and genomic resources from D. melanogaster, to measure the evolution of gene expression patterns between D. sechellia and D. simulans. We are then testing the phenotypic effects of these gene expression changes by constructing transgenic D. simulans.
Research Interests
- The evolution of developmental mechanisms
- Early patterning in molluscs and related groups
- Cytoskeletal basis of asymmetric cell divisions
- Evolution of novel phenotypes
Recent Publications
Longjun Wu, Kailey E Ferger, J David Lambert, Ilya Ruvinsky Gene Expression Does Not Support the Developmental Hourglass Model in Three Animals with Spiralian Development Molecular Biology and Evolution, 2019
Johnson, A.B., Fogel, N.S., Lambert, J.D. Growth and morphogenesis of the gastropod shell Proceedings of the National Academy of Sciences of the United States of America, 2019
Lambert, J.D., Johnson, A.B., Hudson, C.N., Chan, A. Dpp/BMP2-4 Mediates Signaling from the D-Quadrant Organizer in a Spiralian Embryo Current Biology, 2016
Goulding, M.Q., Lambert, J.D. Mollusc models I. The snail Ilyanassa Current Opinion in Genetics and Development, 2016
Chan, X.Y., Lambert, J.D. Development of blastomere clones in the Ilyanassa embryo: Transformation of the spiralian blastula into the larval body plan Development Genes and Evolution, 2014
Gharbiah, M., Nakamoto, A., Johnson, A.B., Lambert, D.J., Nagy, L.M. Ilyanassa Notch signaling implicated in dynamic signaling between all three germ layers International Journal of Developmental Biology, 2014
Chan, X.Y., Lambert, J.D. Patterning a spiralian embryo: A segregated RNA for a Tis11 ortholog is required in the 3a and 3b cells of the Ilyanassa embryo Developmental Biology, 2011
Rabinowitz, J.S., Lambert, J.D. Spiralian quartet developmental potential is regulated by specific localization elements that mediate asymmetric RNA segregation Development, 2010
Lambert, J.D., Chan, X.Y., Spiecker, B., Sweet, H.C. Characterizing the embryonic transcriptome of the snail iljanassa Integrative and Comparative Biology, 2010
Lambert, J.D. Developmental Patterns in Spiralian Embryos Current Biology, 2010
For more, please visit my ORCID Profile
