THESIS in Biology
The effect of modifications to DNA structure on the architecture of bacterial DNA condensed with a nucleoid-associated protein
Fall 2024, Volume 23, Issue 1
Kevin Zheng ’24, Azra Walker, Anne S. Meyer*
https://doi.org/10.47761/JLYC7218
Introduction
Dps serves multiple functions in bacterial cells, but one of its primary functions is binding to and condensing DNA. This function protects the genetic material of the bacteria from environmental stressors, allowing the bacteria to survive in a wide range of harsh conditions. Under both stress and non-stress conditions, Dps monomers are commonly assembled into a spherical dodecamer that is 90 Angstroms in diameter, with a hollow core cavity that is 45 Angstroms in diameter (1). Each Dps monomer consists of a bundle of four alpha-helices interacting with each other (2). Along the surface of the dodecamer, most of the surface is negatively charged, making it unsuitable for interactions with the negatively charged phosphate backbone of DNA molecules (3). Studies suggest that the N-terminal tails of the Dps dodecamer may mediate the interaction with DNA (10). These N-terminal tails are largely disordered and extend outwards from the 12-mer quaternary structure of the Dps dodecamer. In addition, they contain three positively charged lysine residues at the 5, 8, and 10th positions required for DNA binding (4).
The interaction between the lysine residues on the N-terminal tails of the Dps with the DNA occurs when one Dps oligomer interacts with multiple DNA molecules simultaneously. Numerous interactions are possible because the Dps oligomer has multiple N-terminal tails projecting out of it. The size of the tails is small compared to the overall size of the Dps dodecamer, resulting in minimal opportunities for interactions between the residues on the tails and DNA if there is only one strand of DNA. As a result, DNA plectonemes have been shown to have the most stable condensate formation between Dps and DNA (S2 Figure 1A-B). When one DNA strand is twisted around itself, it allows for the distance between neighboring regions of the plectoneme to be roughly the same length as the dodecamer diameter, allowing the Dps dodecamer to interact with both strands (5).
Since electrostatic interactions drive Dps-DNA interactions, it is most likely that the different morphologies observed in Dps-DNA condensate under various environmental conditions are due to changes in these electrostatic interactions (6). Using an in-vitro system and different buffer conditions, we have discovered that the Dps-DNA condensates can take on three different morphologies: globular, liquid-like, and spongiform (S1 Figure 1A-C). When using in-vivo systems where the cells have been deprived of nutrients for an extended period, others have observed additional morphologies of the condensates: nanocrystalline, liquid crystalline, and folded nucleosome-like type (7). These different condensate structures vary in their DNA packing architecture and potential accessibility to DNA molecules.
Despite obtaining a better understanding of the formation of Dps-DNA condensates and how the morphologies of these condensates change under various environmental conditions, it is still not understood how the morphologies of the condensate change when the DNA itself is altered. Upon exposure to Escherichia coli genomic DNA, which is circular and of a specific length by nature, Dps will condense DNA and form a Dps-DNA condensate. However, not all organisms share the same features of DNA, and little is known about how Dps will interact with these different features of DNA that deviate from that of Escherichia coli. Features of DNA that could vary include fragments of various lengths and supercoiling states, as some are circular while others are linear by nature. This study examines the question of how Dps interacts with different features of DNA, which is essential to better understand the function of Dps in organisms besides E. coli and how those organisms respond to environmental stresses.
Materials and Methods
Construction of different lengths of DNA fragments.
Using Lambda phage DNA, two fixed forward primers and multiple reverse primers were designed to create DNA fragments of different lengths (Table 1). The reverse primers were designed to create five DNA fragments of different lengths.These lengths were determined based on the distance between the forward primers and respective reverse primers. One forward primer was used to develop the 500, 1000, and 2000 bp, and the other was used to create DNA fragments of 5000 and 10000 bp in length. Lambda DNA was used as a control, roughly 50,000 bp in length.
We created a master mix consisting of the forward primer, DNA polymerase, the template DNA being Lambda DNA, and other components necessary to synthesize new DNA strands before PCR amplification. After distributing the master mix into five PCR tubes for each of the five different DNA lengths being tested at equal amounts, the reverse primers were added to each of their respective PCR tubes (8). After each DNA fragment was amplified, each group of DNA fragment lengths was purified using a PCR cleanup kit to remove the impurities and components that were not the DNA fragments. A sample from each group of purified DNA fragments was analyzed via gel electrophoresis to ensure they were synthesized at their intended lengths. The resulting DNA bands were compared to a DNA ladder, where each DNA product corresponded correctly to its size (Figure 1A-B).
| Primers | Sequence |
| Forward Primer #1 | GCTGGGTGTTTATGCCTAC |
| 500 Reverse Primer | CGCCAAATTTAAGATACTGCTCC |
| 1000 Reverse Primer | GTCAATCAGCCAGCTTTCC |
| 2000 Reverse Primer | GCATCCACACTTTCACTCG |
| Forward Primer #2 | GCTTGGTGTACCTCATCTAC |
| 5000 Reverse Primer | GTTATCAAGCACTGCACTGG |
| 10000 Reverse Primer | GCTGGCAACTAATTCAGTCC |
Table 1: Sequences for the forward and reverse primers used to create the different lengths of DNA fragments. (A) Forward Primer #1 was used with the 500, 1000, and 2000 reverse primer, while forward primer #2 was used with the 5000 and 10000 reverse primer. (B) All primers satisfy the following requirements: at least 18 bp long, G-C content ranging from 50% to 55%, and having a melting temperature of around 50 degrees Celsius.
Creation of circular and linear DNA fragments.
Three circular plasmids similar to the DNA lengths tested in previous experiments on different-length DNA fragments were identified. Three bacterial strains, each with the plasmid of interest were selected. The three strains separately containing CD8 plasmid (4894 bp), pET17b plasmid (2070 bp), and pShew:mtrC (10,500 bp). LB medium was inoculated with the three bacterial strains along with their respective antibiotics: for the strain containing the CD8 and pShew:mtrC plasmid, the antibiotics used were Kanamycin, while for the strain containing the pET17b plasmid, Chloramphenicol was used. The strains were then shaken at 37 °C overnight to allow for the growth of a saturated culture. After the incubation period, the plasmids of each length were isolated from the bacteria using a plasmid miniprep kit.
A portion of the extracted plasmids of each of the three lengths was then utilized to undergo restriction enzyme digest to create relaxed DNA, (which will be henceforth referred to as linear DNA). Restriction enzymes were chosen based on their ability to only cut the plasmid at one position, thereby creating a linear piece of DNA of that length. If a restriction enzyme cut the plasmids at multiple positions, it would result in numerous linear fragments of much smaller lengths. The CD8 and pShew:mtrC plasmid was cut using the restriction enzyme EcoR1. In contrast, the pET17b plasmid was cut using the restriction enzyme Xbal. In the control group, the CD8 plasmid was incubated with a restriction enzyme for which it has no cut site, so it could not cut the plasmid. After obtaining the relaxed DNA from a portion of each extracted plasmid of different lengths through restriction digest, the resulting linear DNA was purified using a PCR cleanup kit to remove all the impurities. To ensure the plasmids were cut at the length we expected, each circular and linearized plasmid was loaded onto a gel and analyzed via gel electrophoresis. The resulting bands were then compared to a DNA ladder, where each DNA product used corresponded correctly to their respective sizes and location on the gel, with circular plasmids migrating further down the gel than their respective linear counterparts (Figure 1C).
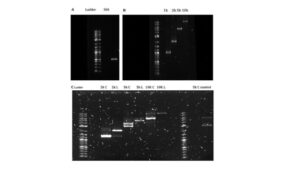 Figure 1. Measurement of the size of purified DNA products of different lengths and supercoiling state relative to the 1 Kb Plus Ladder. (A-B) Gel electrophoresis of the purified DNA products of different lengths after PCR amplification and cleanup. (C) Gel electrophoresis of the purified DNA products of different supercoiling states after selected plasmids of specific lengths were extracted from bacteria, and some undergo a restriction digest. The C represents the circular form of the DNA, while the L represents the linear form of the DNA.
Figure 1. Measurement of the size of purified DNA products of different lengths and supercoiling state relative to the 1 Kb Plus Ladder. (A-B) Gel electrophoresis of the purified DNA products of different lengths after PCR amplification and cleanup. (C) Gel electrophoresis of the purified DNA products of different supercoiling states after selected plasmids of specific lengths were extracted from bacteria, and some undergo a restriction digest. The C represents the circular form of the DNA, while the L represents the linear form of the DNA.
Image Preparation and Imaging using Fluorescence Microscopy.
Nine samples were made to assemble the Dps-DNA complexes for imaging. Each sample contained the same buffer containing 5% polyethylene glycol 8k (PEG), 4 mM MgCl2, 50 mM HEPES-KOH, and 100 mM KCl. Then, we added 1x sytox green and 10 ng/uL of the aforementioned DNA fragments (final concentrations). Finally, 1 mM Dps was added to begin forming condensates. We made three control groups: a sample with all components but DNA, another with all components but Dps, and the final sample containing neither Dps nor DNA. We ensured that the amount of DNA was consistent between samples. The use of sytox green allows for the visualization of DNA. Some of the Dps we used is a variant chemically labeled with Alexa fluor 647, enabling the Dps to be visualized. Most, however, were wild-type Dps (9). All samples were incubated overnight at 37 °C and imaged the following day using fluorescent microscopy.
Following the same procedure described above, nine samples were made to assemble the Dps-DNA complexes for imaging. Each sample had the same buffer at the same concentration described above. The sole differences were the lengths of DNA fragments, structure of DNA (circular or linear), and the final DNA concentration. Two samples shared the same size and concentration of DNA fragments but differed in the DNA topology as one consisted of supercoiled DNA, while the other sample consisted of relaxed DNA. This setup was applied to three DNA fragment lengths (2000, 5000, and 10000-bp).
The three control samples included a mock-digested 5000-bp plasmid incubated with EcoR1(a restriction enzyme for which it has no cut sites), a sample with no DNA, and a sample with neither DNA nor Dps. The samples were incubated overnight at 37 °C. The following day, the samples were imaged using fluorescence microscopy. For each sample, images of the condensates were taken at five random positions. Bright-field and green-fluorescent images were used for each position. This procedure was done for three trials.
Data Analysis using software.
The green images of the condensates, representing the DNA, were analyzed using a custom Fiji program that detects fluorescence distribution throughout the surface of the Dps-DNA condensates. Fiji sums the intensity distributed throughout a condensate, referred to as integrated density (IntDen), as well as the area and perimeter of the Dps-DNA condensate. Data regarding area and perimeter was used to calculate the circularity of each condensate formation. The formula for circularity that was used was:
 The 12.56 represents 4 times pi, which should equal 1 if the shape is a perfect circle when multiplied by this value. The distance from 1 of this value is commonly used to describe circularity.
The 12.56 represents 4 times pi, which should equal 1 if the shape is a perfect circle when multiplied by this value. The distance from 1 of this value is commonly used to describe circularity.
Results
Binding affinity and cooperativity differences between Dps and circular and linear DNA remain statistically insignificant.
Dps, a protein commonly found in bacteria, typically binds to circular DNA. However, the affinity of Dps for linear DNA has yet to be tested. To test this affinity, we performed a gel shift assay. The bands will appear at similar concentrations if Dps binds to linear or circular DNA with the same affinity.
No binding was observed between Dps and DNA when 0.1 μM Dps was present. The well containing 0.3 μM Dps displayed binding, indicating that the concentration required for Dps to bind to circular is within the 0.1 to 0.3 μM range. The mock digest and linearized plasmid indicate comparable binding affinity (Figure 2A-B).
We calculated the dissociation constant (KD) and Hill coefficient by creating a binding curve. The mock digest and circular DNA fragment shared similar values, with the KD being roughly 0.25 (Figure 2C). The linear DNA fragments had a slightly lower KD value of around 0.23; the slightly lower KD value between Dps and linear DNA fragments indicates that Dps has a slightly higher binding affinity for linear DNA fragments than circular DNA fragments (Figure 2D). However, the difference is statistically insignificant, and does not conform to expectation, as we would expect Dps to bind to circular DNA at a similar or higher affinity. All experimental groups displayed a Hill coefficient greater than 1, indicating that Dps exhibits positive cooperativity whether interacting with circular or linear DNA fragments (Figure 2D).
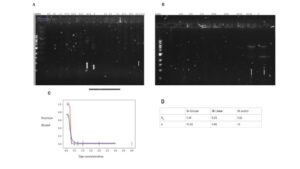 Figure 2: Dissociation and Binding Cooperativity between Dps and DNA fragments of different supercoiling states. Data provided by Azra Walker. (A-B) Gel shift displaying the concentration of Dps needed to bind to DNA. The category of the wells can be read as the (concentration of Dps – whether it is circular (C), linear (L), or control group (CC)). (C )The resulting binding curve was obtained using data from the gel shift (D). The dissociation constant and Hill coefficient values were used for the interaction between Dps with the control group, circular DNA fragment, and linear DNA fragment of 5000 in length.
Figure 2: Dissociation and Binding Cooperativity between Dps and DNA fragments of different supercoiling states. Data provided by Azra Walker. (A-B) Gel shift displaying the concentration of Dps needed to bind to DNA. The category of the wells can be read as the (concentration of Dps – whether it is circular (C), linear (L), or control group (CC)). (C )The resulting binding curve was obtained using data from the gel shift (D). The dissociation constant and Hill coefficient values were used for the interaction between Dps with the control group, circular DNA fragment, and linear DNA fragment of 5000 in length.
The morphologies of the Dps-DNA condensate remain consistent when changing the length of the DNA fragments.
To test the impact of DNA length on the size and morphology of Dps, DNA condensates were created in vitro and imaged using fluorescence microscopy. By analyzing these images, condensate morphology and size could be approximated using the summed intensity of DNA fluorescence across them. Condensates share the same spongiform morphology regardless of DNA fragment size (Figure 3A-F). This suggests that changing the DNA length does not alter the electrostatic interaction between the Dps and DNA (11) as it was previously discussed that this was one of the main driving forces in forming the condensate morphology (6).
 Figure 3. Dps-DNA condensates adopt spongiform morphologies regardless of differences in DNA fragment lengths. (A-F) The Dps-DNA condensates formed when Dps is incubated with DNA fragments of different lengths.
Figure 3. Dps-DNA condensates adopt spongiform morphologies regardless of differences in DNA fragment lengths. (A-F) The Dps-DNA condensates formed when Dps is incubated with DNA fragments of different lengths.
There is a marked increase in condensate size when formed using intermediate DNA lengths.
While little appears to change morphologically in Dps-DNA condensates when using longer or shorter DNA fragments, there are differences in the level of total fluorescence (representing DNA) inside these condensates. This indicates that altering the DNA’s size affected condensate formation but was unrelated to its morphology. The average summed intensity of DNA fluorescence in each condensate was used to represent the average condensate size. Through this analysis, Dps-DNA condensates were observed to be, on average, larger when Dps was incubated with 5000-bp DNA fragments compared to other DNA lengths (Figure 4A-F). This suggests that changing the size of the DNA may not alter the electrostatic interactions between DNA and Dps (7) but could still change the level of accessibility between the DNA fragments and Dps. This in turn changes how frequently Dps will find and bind to DNA (5).
 Figure 4: Fluorescent images of the DNA in condensates formed using different lengths of DNA. (A-F) Distribution of fluorescence of the Dps-DNA condensate formed from incubating Dps with different lengths of DNA fragments. The level of fluorescence indicates the DNA present in each of the condensates.
Figure 4: Fluorescent images of the DNA in condensates formed using different lengths of DNA. (A-F) Distribution of fluorescence of the Dps-DNA condensate formed from incubating Dps with different lengths of DNA fragments. The level of fluorescence indicates the DNA present in each of the condensates.
Optimal DNA fragment lengths for Dps-DNA condensate of the largest size and area.
In all three trials, after taking the average integrated density of all the condensates visible in each image, there was a prominent peak in the average integrated density of the Dps-DNA condensate formed from 5000-bp DNA fragments (Figure 5B). This peak suggests that when incubated with 5000-bp DNA fragments, it alters the DNA in a manner that changes the number of binding opportunities Dps can have when in contact with DNA fragments of this length. In addition, all three trials showed the integrated density value of the condensate formed when Dps is incubated with DNA fragments that are shorter and longer than 5000-bp being drastically smaller than the values of that when the condensate is formed with 5000-bp DNA fragments. These lower values indicate a significantly smaller condensate (Figure 5B).
When comparing the average area of the Dps-DNA condensate across all the different DNA fragment lengths used, it can be visibly seen that the average area value for condensates formed when Dps is incubated with 5000- bp DNA fragment was larger than that of other groups (Figure 5A). This indicates that these condensates have larger surfaces, supporting our hypothesis that they are the largest. However, it is essential to note that the error bar does overlap, signifying that there is no significant difference in the area among condensates formed when Dps is incubated with 2000, 5000, and 10000-bp DNA fragments.When comparing the average circularity, there is no significant difference in the circularity of the condensates formed when incubating Dps with different DNA fragment lengths (Figure 5C).
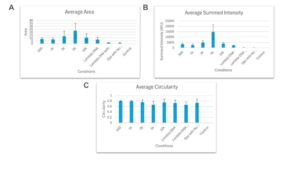 Figure 5: A graph displaying the average area, summed intensity, and circularity of Dps-DNA condensates present in samples using specific DNA lengths. (A) Average area of the Dps-DNA condensates formed from each of the different DNA fragment lengths used (B) Average IntDen of the Dps-DNA condensates formed from using different lengths of DNA fragment (C) Average Circularity of the Dps-DNA condensates formed from using different DNA fragment lengths
Figure 5: A graph displaying the average area, summed intensity, and circularity of Dps-DNA condensates present in samples using specific DNA lengths. (A) Average area of the Dps-DNA condensates formed from each of the different DNA fragment lengths used (B) Average IntDen of the Dps-DNA condensates formed from using different lengths of DNA fragment (C) Average Circularity of the Dps-DNA condensates formed from using different DNA fragment lengths
No significant change in size or structure of Dps-DNA condensate when under different supercoiled states
Samples of DNA molecules of the same size but different topologies were prepared to determine whether DNA supercoiling would affect the morphology and characteristics of resulting Dps-DNA condensates. The fluorescent images of these samples were then analyzed in the same manner as previous images.
Similar to how varying lengths of DNA did not affect the overall morphology of Dps-DNA condensates, the state of DNA molecules (supercoiled or relaxed) had no observable effect on the structure of the condensates, as all condensates took on a spongiform morphology (Figure 6A-6G). When comparing the total average summed integrated density value, there was no significant difference in the size of condensates formed with supercoiled DNA compared to condensates formed with relaxed DNA. Condensates formed when Dps were incubated with 2000-bp circular DNA plasmids were roughly the same size when formed with 2000-bp linear DNA plasmids. The same is true of 5000 and10000-bp plasmids, regardless of whether it was supercoiled or relaxed. (Figure 6I).
There was no significant difference in the total area and circularity among the condensates formed from the circular and linear DNA fragments of varying fragment lengths (Figure 6H and J). This further manifests the lack of notable difference between condensates formed with different supercoiling states. Condensates formed from both the mock-digested 5000-bp plasmid and 5000-bp circular DNA fragments appeared to be relatively similar (Figure 6G). Overall, no significant differences were noted across all experiments comparing condensate formation using same-size DNA molecules, but they differed in supercoiling states (Figure 6A-G).
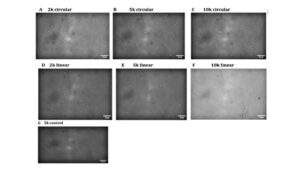
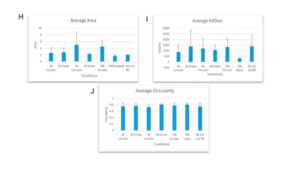 Figure 6: Condensates formation when Dps is incubated with DNA of different lengths and whether DNA is supercoiled or not. (A-G) Condensate formation when Dps is incubated with DNA molecules that share the same length but different DNA topology (H) Average area for the Dps-DNA condensates formed under different DNA length and supercoiling states (I) Average IntDen of the Dps-DNA condensates formed under different DNA length and supercoiling states (J) Average Circularity of the Dps-DNA condensates formed under different DNA length and supercoiling states
Figure 6: Condensates formation when Dps is incubated with DNA of different lengths and whether DNA is supercoiled or not. (A-G) Condensate formation when Dps is incubated with DNA molecules that share the same length but different DNA topology (H) Average area for the Dps-DNA condensates formed under different DNA length and supercoiling states (I) Average IntDen of the Dps-DNA condensates formed under different DNA length and supercoiling states (J) Average Circularity of the Dps-DNA condensates formed under different DNA length and supercoiling states
Discussion
The findings show that changing the DNA length at our concentration affects the size and number of condensates, without changing the overall structure. This suggests that there is likely some difference in the ability to form complex structures between smaller and larger DNA fragments, which affects the capacity of Dps to bind to these DNA. Samples with larger DNA fragments may be more accessible for Dps to attach to and condense, creating smaller structures. Samples with smaller DNA fragments could condense more effectively up to a point. With more free DNA ends, less DNA needs to wrap around the Dps for condensation to occur (11). The lesser need for DNA bending would mean that it is faster and more efficient for small DNA fragments to wrap themselves around the Dps completely.
DNA fragments smaller than 5000 bp in length have more free DNA ends, helping them condense more easily by the Dps, and DNA fragments larger than 5000 bp in length are more accessible for Dps to attach and condense. DNA fragments that are 5000-bp in length are at a point between where these two effects occur. Our data suggests that DNA fragments of 5000-bp in length are either the most difficult to condense, condense less effectively when done, or both. DNA fragments of 5000-bp may condense less effectively and are harder to condense due to their limited length. Small DNA fragments can result in more effective condensing. Small DNA is short enough that its free DNA end can wrap around the Dps dodecamer without bending. This makes it easier for an array of small DNA fragments to line up, which in turn promotes interactions between the Dps dodecamer and multiple DNA fragments simultaneously. It is thus more likely for all the N-terminus tails in the Dps dodecamer to interact with the DNA. More interaction leads to a higher level of condensing.
Medium-length DNA fragments, like the 5000-bp fragment have ends that aren’t long enough to bend completely, unlike longer-length DNA fragments where the free DNA ends are long enough to bend completely to reach the Dps that are already interacting with multiple DNA strands. As a result, the inability to bend completely back will cause the DNA fragment to bend instead, making it harder for the DNA region to be accessible to some sites of the Dps dodecamer., This reduces the number of interactions between Dps and DNA, resulting in less wrapping and less condensing.
Small differences were observed in the overall size of the Dps-DNA condensate between the condensates formed when Dps was incubated with circular plasmids, and the condensates formed when Dps was incubated with linear plasmids. However, the data does not display a significant difference between the sizes of the condensates, indicating that Dps can condense both linear and circular plasmids in roughly the same manner and form. Similar to how the Dps interacts with DNA fragments of different lengths, the overall morphology of the Dps-DNA condensate remains consistently spongiform.
Whether the DNA is in a supercoiled state did not impact the resulting condensate’s overall size or structure greatly. This is most likely because supercoiled DNA contains multiple DNA regions interwound that were initially distal to each other (12). Similar to how medium-sized linear DNA fragments may bend in a manner that makes it harder to be accessible to some sites of the Dps dodecamer, distal DNA regions that are interwound multiple times, potentially limit the accessibility of DNA to Dps. As a result, Dps can only interact with DNA in certain regions. With most of the DNA already intertwined with each other, very few DNA will wrap around the Dps, resulting in bigger or similar size condensates as those formed with medium-sized DNA fragments.
Recent research has provided a more in-depth understanding of the structure of Dps itself, how it interacts with DNA to protect it, the different structures the Dps-DNA condensate can take on, and how changing environmental conditions could alter these structures. Although studies have focused on how changes to the environment have affected the interaction between Dps and DNA, there has been minimal research on whether altering one of the key components of the condensate DNA, would change its overall morphology. However, our results have indicated that altering the DNA lengths and supercoiling states does not affect the overall morphology of Dps-DNA condensates. Still, it alters the interaction between the DNA and Dps in a manner that changes some condensate characteristics. These results could have certain implications for non-bacterial organisms. Not all organisms share the same structure and length of DNA as E. coli, which possess circular and significantly longer DNA than other bacteria. Observing how Dps binds to and interacts with these different DNA modifications y helps provide a better understanding of the possible function of Dps in other organisms and environmental conditions. . A complete understanding of the role of Dps in other organisms would require extensive testing of additional DNA modifications, such as methylated DNA, and their effect on Dps-DNA interactions.
Supplementary Information
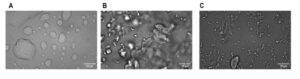 S1 Figure 1: Three different morphologies of Dps-DNA condensate. (A-C) Azra Walker provided images where three different Dps-DNA condensate morphologies were discovered in the Meyer Lab: Liquid-like, globular, and spongiform.
S1 Figure 1: Three different morphologies of Dps-DNA condensate. (A-C) Azra Walker provided images where three different Dps-DNA condensate morphologies were discovered in the Meyer Lab: Liquid-like, globular, and spongiform.
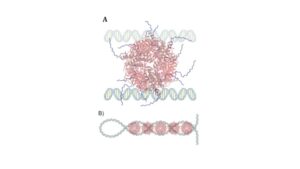 S2 Figure 1: The model depicts DNA Plectonemes as one possible interaction between Dps and DNA (11). (A) Dps interacting with two parallel linear DNA strands (B) Dps interacting with a supercoiled or interwound DNA strand.
S2 Figure 1: The model depicts DNA Plectonemes as one possible interaction between Dps and DNA (11). (A) Dps interacting with two parallel linear DNA strands (B) Dps interacting with a supercoiled or interwound DNA strand.
References
- Ceci P, Cellai S, Falvo E, Rivetti C, Rossi GL, Chiancone E. DNA condensation and self-aggregation of Escherichia coli Dps are coupled phenomena related to the properties of the N-terminus. Nucleic Acids Res. 2004 Nov 8;32(19):5935-44. doi: 10.1093/nar/gkh915. PMID: 15534364; PMCID: PMC528800.
- Garibyan L, Avashia N. Polymerase chain reaction. J Invest Dermatol. 2013 Mar;133(3):1-4. doi: 10.1038/jid.2013.1. PMID: 23399825; PMCID: PMC4102308.
- Grant RA, Filman DJ, Finkel SE, Kolter R, Hogle JM. The crystal structure of Dps, a ferritin homolog that binds and protects DNA. Nat Struct Biol. 1998 Apr;5(4):294-303. doi: 10.1038/nsb0498-294. PMID: 9546221.
- Harteis S, Schneider S. Making the bend: DNA tertiary structure and protein-DNA interactions. Int J Mol Sci. 2014 Jul 14;15(7):12335-63. doi: 10.3390/ijms150712335. PMID: 25026169; PMCID: PMC4139847.
- Karas VO, Westerlaken I, Meyer AS. The DNA-Binding Protein from Starved Cells (Dps) Utilizes Dual Functions To Defend Cells against Multiple Stresses. J Bacteriol. 2015 Oct;197(19):3206-15. doi: 10.1128/JB.00475-15. Epub 2015 Jul 27. PMID: 26216848; PMCID: PMC4560292.
- Ma J, Wang MD. DNA supercoiling during transcription. Biophys Rev. 2016 Nov;8(Suppl 1):75-87. doi: 10.1007/s12551-016-0215-9. Epub 2016 Jul 13. PMID: 28275417; PMCID: PMC5338639.
- Melekhov VV, Shvyreva US, Timchenko AA, Tutukina MN, Preobrazhenskaya EV, Burkova DV, Artiukhov VG, Ozoline ON, Antipov SS. Modes of Escherichia coli Dps Interaction with DNA as Revealed by Atomic Force Microscopy. PLoS One. 2015 May 15;10(5):e0126504. doi: 10.1371/journal.pone.0126504. PMID: 25978038; PMCID: PMC4433220.
- Loiko N, Danilova Y, Moiseenko A, Kovalenko V, Tereshkina K, et al. (2020) Morphological peculiarities of the DNA-protein complexes in starved Escherichia coli cells. PLOS ONE 15(10): e0231562. https://doi.org/10.1371/journal.pone.0231562
- Orban K, Finkel SE. Dps Is a Universally Conserved Dual-Action DNA-Binding and Ferritin Protein. J Bacteriol. 2022 May 17;204(5):e0003622. doi: 10.1128/jb.00036-22. Epub 2022 Apr 5. PMID: 35380871; PMCID: PMC9112962.
- Roy S, Saraswathi R, Gupta S, Sekar K, Chatterji D, Vijayan M. Role of N and C-terminal tails in DNA binding and assembly in Dps: structural studies of Mycobacterium smegmatis Dps deletion mutants. J Mol Biol. 2007 Jul 20;370(4):752-67. doi: 10.1016/j.jmb.2007.05.004. Epub 2007 May 10. PMID: 17543333.
- Schirripa Spagnolo C, Luin S. Choosing the Probe for Single-Molecule Fluorescence Microscopy. Int J Mol Sci. 2022 Nov 29;23(23):14949. doi: 10.3390/ijms232314949. PMID: 36499276; PMCID: PMC9735909.
- Shahu S, Vtyurina N, Das M, Meyer AS, Ganji M, Abbondanzieri EA. Bridging DNA contacts allow Dps from E. coli to condense DNA. bioRxiv [Preprint]. 2024 Jan 25:2024.01.22.576774. doi: 10.1101/2024.01.22.576774. Update in: Nucleic Acids Res. 2024 Apr 04;: PMID: 38328146; PMCID: PMC10849575.
About the Author
Kevin Zheng recently graduated in May 2024, earning a BS in Biochemistry with a minor in psychology. He started conducting research at the Meyer Lab in the fall of 2022 and contributed to the project involving the effect of different DNA features on the formation of DNA-Dps condensate up until graduation. His undergraduate research interest focuses on better understanding the function of Dps in various organisms. Currently, Kevin is pursuing an EdM at the Harvard Graduate School of Education, where his research examines the impact that mentorship has on the likelihood of undergraduate students pursuing teaching as a postgraduate career.
Cite this Article
Zheng, K., Walker, A., Meyer A. S. (2024). The effect of modifications to DNA structure on the architecture of bacterial DNA condensed with a nucleoid-associated protein. University of Rochester, Journal of Undergraduate Research, 23(1). https://doi.org/10.47761/JLYC7218
JUR | Creative Commons Attribution 4.0 BY International License
