|
|
|
|
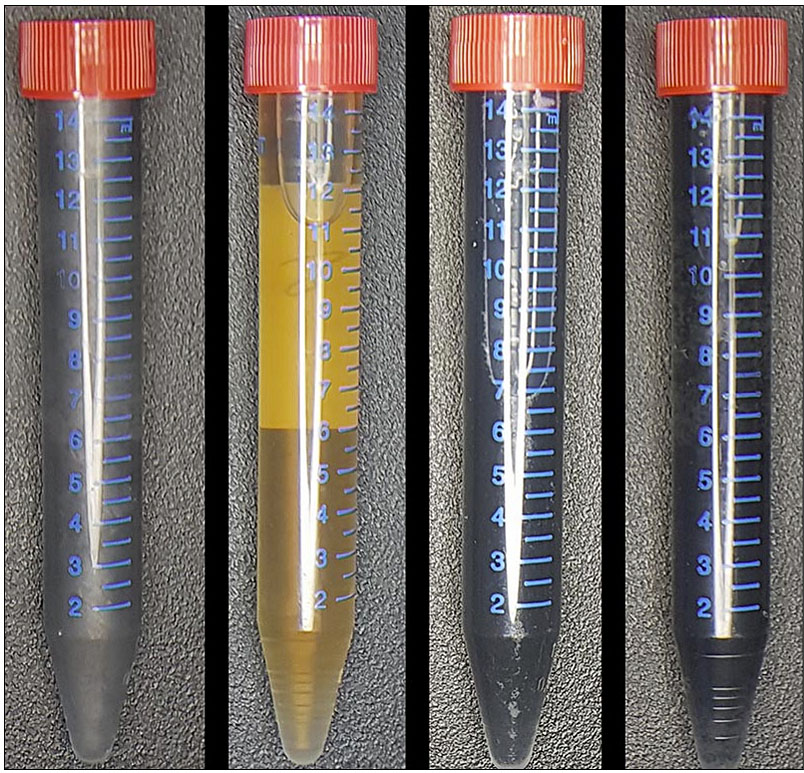 From left to right, a vial of graphite (Gr), like you would find in an ordinary pencil; a vial of graphene oxide (GO), produced by exfoliating Gr—shedding the layers of the material—and mixing it with the bacteria Shewanella; a vial of the resulting product—graphene materials (mrGO); and a vial of graphene materials that have been produced chemically (crGO). The graphene materials produced by Anne Meyer’s lab are significantly thinner than the graphene materials produced chemically. (Delft University of Technology photo / Benjamin Lehner)
Will your future computer be made using bacteria?Graphene—a flake of carbon as thin as a single layer of atoms—is a revolutionary nanomaterial due to its ability to easily conduct electricity, as well as its extraordinary mechanical strength and flexibility. However, a major hurdle in adopting it for everyday applications is producing graphene at a large scale, while still retaining its amazing properties.
In a paper published in the journal ChemOpen, Anne S. Meyer, an associate professor of biology, and her colleagues at Delft University of Technology in the Netherlands describe a novel technique to overcome this barrier: mixing oxidized graphite with bacteria. Their method is a more cost-efficient, time-saving, and environmentally friendly way of producing graphene materials versus those produced chemically, and could lead to the creation of innovative computer technologies and medical equipment.
Graphene is extracted from graphite, the material found in an ordinary pencil. At exactly one atom thick, graphene is the thinnest—yet strongest—two-dimensional material known to researchers. Scientists from the University of Manchester in the United Kingdom were awarded the 2010 Nobel Prize in Physics for their discovery of graphene; however, their method of using sticky tape to make graphene yielded only small amounts of the material.
In order to produce larger quantities, Meyer and her colleagues started with a vial of graphite. They exfoliated the graphite—shedding the layers of material—to produce graphene oxide (GO), which they then mixed with the bacteria Shewanella. They let the beaker of bacteria and precursor materials sit overnight, during which time the bacteria reduced the GO to a graphene material.
The bacterially-produced graphene material created in Meyer’s lab is not only conductive, but also thinner and more stable than graphene produced chemically. Additionally, it can be stored for longer periods of time, making it well suited for a variety of applications, including field-effect transistor (FET) biosensors and conducting ink.
Read more here.
Aluie, Matson recognized by DOE as exceptional researchersTwo University faculty members – Hussein Aluie and Ellen Matson – have been named recipients of Early Career Research awards from the Department of Energy (DOE). The award, now in its 10th year, is designed to bolster the nation’s scientific workforce by providing support to exceptional researchers during the crucial early career years, when many scientists do their most formative work.
Matson and Aluie are among 73 scientists – including 27 from DOE’s national laboratories and 46 from U.S. universities – to receive the award from DOE’s Office of Science this year. For university faculty members, the awards pay $150,000 a year for five years.
Aluie, an assistant professor of mechanical engineering, will use a new method his group has been developing to advance numerical modeling of the incredibly complex fluid instabilities that hinder achieving controlled nuclear fusion in the lab. A self-sustaining fusion reaction could provide a virtually endless supply of clean energy.
However, when lasers converge on the exterior of a deuterium/tritium fuel capsule during an inertial confinement fusion experiment at the Laboratory for Laser Energetics, the lasers turn the outer shell into a hot, low-density plasma. This plasma pushes against the colder, denser shell inside. Different materials start inter-penetrating through each other, like fingers, removing heat from the center and bringing in impurities from the outside. The target’s core fails to reach sufficiently high temperatures and densities. Numerical models that attempt to predict these fluid instabilities and mixing fall short because they fail to sufficiently resolve the vorticity (local spinning) at various locations within the plasma and are unlikely to be resolved in the near future due to exorbitant computing costs.
Aluie seeks to address this challenge by developing a nonlinear mixing model capable of accounting for the missing vorticity. The research plan includes extensive development, testing, and validation. “The project should enable HED (high-energy density) application codes, such as those used in fusion, to better predict instability growth and mixing at a significantly lower computational cost,” Aluie says. The project may also prove valuable beyond HED applications, such as in astrophysics, he says.
Matson, an assistant professor of chemistry, will use her award to expand her research program into heavy element chemistry. In particular, the laboratory will begin investigating the electronic communication between four electron-rich actinide elements (thorium, uranium, neptunium and plutonium) and metal-oxide clusters.
The emphasis of this work rests in understanding the way these elements engage in sharing electrons with electron-poor metal-oxide assemblies. This could lead to new reactivity pathways for these radioactive atoms, and also new applications in separations chemistry. In particular, her work could lead to the development of well-defined, chemical uses for the long-lived radionuclides of the nuclear fuel cycle – including both the products of front-end enrichment processes and the back-end spent fuels.
The award allows Matson to return to her scientific roots. Matson obtained her PhD in chemistry studying the synthesis, reactivity and electronic structure of electron-rich, uranium-carbon bonds. These new projects will merge Matson’s interests in multimetallic assemblies with her love of the actinide elements.
Click here to read abstracts of this year’s winning projects.
Med Center researchers provide details of Batten diseaseAn article in Neurology Today describes how Medical Center (URMC) researchers have painstakingly compiled decades of patient data and developed a highly sensitive rating scale that provides a detailed picture of Batten disease. This tool, which was developed 17 years ago, has set a standard for how to conduct natural history research in rare childhood neurodegenerative diseases.
A common challenge in the treatment and study of neurological disorders is that these diseases are often poorly understood. The complex manifestation of these conditions – in which the appearance, severity, and progression of symptoms can vary widely – combined with the difficulty in recruiting study participants with rare neurological disorders often conspire to hamper efforts to precisely define the disease. This is a major problem when it comes to clinical trials, where rigorously defined outcome measures are required to determine if an experimental treatment is effective.
CLN3 disease is an example. The disorder is one of a family of conditions called neuronal ceroid lipofuscinoses (NCL), more commonly referred to as Batten disease, which are characterized by vision loss, movement disorders, seizures, and dementia. It is estimated 2 to 4 out of every 100,000 children in the U.S. have NCL. CLN3 disease, a juvenile onset form of NCL, is the most prevalent form of the disease.
Initiated in 2001 by Medical Center neurologists Frederick Marshall and Jonathan Mink, the Unified Batten Disease Rating Scale (UBDRS) includes physical, seizure, behavioral, and vision assessments. Over time, the scale has been refined based on clinical observations. To date, University of Rochester Batten Disease researchers have used the UBDRS to perform almost 500 evaluations in more than 200 patients in the U.S. and around the world.
The development of the UBDRS was followed by the creation of a registry of known cases and the formation of the University of Rochester Batten Center in 2005. The center is co-directed by Mink and Erika Augustine and includes Heather Adams and Amy Vierhile on the leadership team. It has become a leading center internationally for clinical research on all forms of Batten disease.
The traits of a translational scientistTranslational science is a field of investigation that seeks to understand each step in the process of turning observations in the laboratory, clinic and community into interventions that improve the health of individuals and the public. Recently, an international group of investigators who are part of the Translation Together consortium defined the ideal characteristics of a translational scientist. The seven characteristics are:
- Boundary crosser: breaks down silos to collaborate broadly.
- Domain expert: has deep disciplinary expertise in one or more of the translational phases.
- Team player: recognizes and engages in team science to understand and advance translation.
- Process innovator: seeks to understand and improve processes underlying the transitions from one translational phase to another.
- Skilled communicator: can “translate” research effectively to stakeholders across the various domains of translation.
- Systems thinker: can analyze and navigate the internal and external dynamics that impact the translational process.
- Rigorous researcher: adheres to high standards of rigor, reproducibility and transparency.
Read the full article.
Seminar on Illumina technologyA seminar hosted by the Medical Center’s Shared Resource Laboratories will discuss how Illumina technology can help advance research.
The Illumina Next-Generation Sequencing seminar, from 9:15 a.m. to 2 p.m. August 13 at the Hilton Garden Inn at College Town, will feature Illumina experts who will present an overview of next-generation sequencing and discuss the basic concepts and key advantages over traditional technologies.
Speakers include several associates from URMC. Learn more and register.
Upcoming PhD dissertation defenseAstrid Parsons, chemistry, “Transition-Metal-Catalyzed Reactions: Harnessing Metals to Facilitate Organic Transformations.” 10 a.m. August 12, 2019. 1400 Wegmans Hall. Advisor: William Jones.
Mark your calendarToday: “Navigating Deaf and Hearing Collaborations in Science.” Second biennial Deaf-Engaged Academic Forum (DEAF-ROC Conference). Keynote speakers, short talks, breakout sessions, panel discussion and a poster session highlighting the research carried out by undergraduate and graduate students, postdoctoral fellows, faculty, and staff attending the conference. Register for the conference (for free).
Today: Demo session on the new Embark technology that will streamline clinical trial management and allow URMC research teams to reach their full potential. All URMC clinical research faculty and staff are encouraged to attend one or more sessions. 2-3 p.m., Helen Wood Hall, Room 1W-502. To RSVP and request special accommodations, contact embark@urmc.rochester.edu.
Aug. 13: Illumina Next-Generation Sequencing seminar, hosted by the Medical Center’s Shared Resource Laboratories. Illumina experts present an overview of next-generation sequencing, basic concepts, and key advantages over traditional technologies. 9:15 a.m. to 2 p.m. Hilton Garden Inn at College Town. Learn more and register.
Aug. 16: Deadline to submit initial applications to the Environmental Health Sciences Center for pilot project funding of up to $30,000 for one year. Submit applications to Pat Noonan-Sullivan. Contact Deborah Cory-Slechta or Pat Noonan-Sullivan for questions. Learn more.
Aug. 21: Training workshop for TriNetX cohort discovery tool that will replace the current i2b2 tool. With TriNetX investigators can search a limited set of electronic medical record data to determine the feasibility of their clinical trials. Participants should have a basic understanding of the TriNetX interface and be able to build simple queries. 2-4 p.m. SRB 3434A/3434B. Register now. Learn more about TriNetX here.
Aug. 30: Entry forms due for a new annual student competition promoting awareness of Tobacco Regulatory Science. Each team (either an individual or up to four students) develops and presents a proposed solution — a well-developed and researched thought experiment — to address one or more identified Tobacco Regulatory Research Priorities. Winning team will travel to DC/MD to present their solution to the FDA and NIH. Read more here.
Sept. 11: Deadline for students, junior faculty, early career researchers, and entrepreneurs to apply for this year’s Falling Walls competition. Complete the online application here. By Friday, September 27 email a three-panel slide presentation. Then pitch your innovative idea — highlighting a breakthrough that creates a positive impact on science and society — in just three minutes on October 8 at Feldman Ballroom. The winner will represent the University at the International Falling Walls Finale in Berlin. Contact Adele Coelho for questions.
Sept. 13: Symposium on novel applications of technology to accelerate the development of new therapeutics for patients with rare neurological disorders. Hosted by the University and sponsored by the Clinical and Translational Science Institute (UR CTSI). Featuring industry leaders in rare diseases, health technologies and clinical trials. Registration information and agenda available on the TRNDS website.
Sept. 17: Training workshop for TriNetX cohort discovery tool that will replace the current i2b2 tool. With TriNetX investigators can search a limited set of electronic medical record data to determine the feasibility of their clinical trials. Participants should have a basic understanding of the TriNetX interface and be able to build simple queries. 2-4 p.m. SRB 1404. Register now. Learn more about TriNetX here.
Oct. 16: Training workshop for TriNetX cohort discovery tool that will replace the current i2b2 tool. With TriNetX investigators can search a limited set of electronic medical record data to determine the feasibility of their clinical trials. Participants should have a basic understanding of the TriNetX interface and be able to build simple queries. 2-4 p.m. SRB 1416. Register now. Learn more about TriNetX here.
Oct. 22: Deadline to apply for AS&E PumpPrimer II seed funding, typically up to $50,000 for up to one year, for innovative research projects to help the applicant establish a novel research direction and secure extramural funding. Submit proposal via the application portal. All eligibility criterion is enumerated in the guidelines Direct questions to your respective AS&E assistant dean: Arts and Sciences – Debra Haring, debra.haring@rochester.edu; Engineering – Cindy Gary, cindy.gary@rochester.edu.
Nov. 20: Training workshop for TriNetX cohort discovery tool that will replace the current i2b2 tool. With TriNetX investigators can search a limited set of electronic medical record data to determine the feasibility of their clinical trials. Participants should have a basic understanding of the TriNetX interface and be able to build simple queries. 2-4 p.m. SRB 1412. Register now. Learn more about TriNetX here.
|
|
|
|
|